View on TensorFlow.org View on TensorFlow.org
|
 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
 Download notebook Download notebook
|
Overview
TensorFlow implements a subset of the NumPy API, available as tf.experimental.numpy. This allows running NumPy code, accelerated by TensorFlow, while also allowing access to all of TensorFlow's APIs.
Setup
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import tensorflow.experimental.numpy as tnp
import timeit
print("Using TensorFlow version %s" % tf.__version__)
2024-08-15 01:31:55.452313: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:485] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered 2024-08-15 01:31:55.473711: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:8454] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered 2024-08-15 01:31:55.480014: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1452] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered Using TensorFlow version 2.17.0
Enabling NumPy behavior
In order to use tnp as NumPy, enable NumPy behavior for TensorFlow:
tnp.experimental_enable_numpy_behavior()
This call enables type promotion in TensorFlow and also changes type inference, when converting literals to tensors, to more strictly follow the NumPy standard.
TensorFlow NumPy ND array
An instance of tf.experimental.numpy.ndarray, called ND Array, represents a multidimensional dense array of a given dtype placed on a certain device. It is an alias to tf.Tensor. Check out the ND array class for useful methods like ndarray.T, ndarray.reshape, ndarray.ravel and others.
First create an ND array object, and then invoke different methods.
# Create an ND array and check out different attributes.
ones = tnp.ones([5, 3], dtype=tnp.float32)
print("Created ND array with shape = %s, rank = %s, "
"dtype = %s on device = %s\n" % (
ones.shape, ones.ndim, ones.dtype, ones.device))
# `ndarray` is just an alias to `tf.Tensor`.
print("Is `ones` an instance of tf.Tensor: %s\n" % isinstance(ones, tf.Tensor))
# Try commonly used member functions.
print("ndarray.T has shape %s" % str(ones.T.shape))
print("narray.reshape(-1) has shape %s" % ones.reshape(-1).shape)
Created ND array with shape = (5, 3), rank = 2, dtype = <dtype: 'float32'> on device = /job:localhost/replica:0/task:0/device:GPU:0 Is `ones` an instance of tf.Tensor: True ndarray.T has shape (3, 5) narray.reshape(-1) has shape (15,) WARNING: All log messages before absl::InitializeLog() is called are written to STDERR I0000 00:00:1723685517.895102 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.898954 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.902591 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.906282 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.918004 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.921334 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.924792 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.928195 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.931648 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.935182 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.938672 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685517.942206 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.174780 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.176882 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.178882 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.180984 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.183497 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.185413 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.187391 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.189496 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.191405 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.193373 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.195387 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.197409 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.235845 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.237893 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.239848 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.241962 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.243923 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.245869 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.247783 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.249797 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.251701 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.254112 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.256530 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355 I0000 00:00:1723685519.258985 23752 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
Type promotion
There are 4 options for type promotion in TensorFlow.
- By default, TensorFlow raises errors instead of promoting types for mixed type operations.
- Running
tf.numpy.experimental_enable_numpy_behavior()switches TensorFlow to useNumPytype promotion rules (described below). - After TensorFlow 2.15, there are two new options (refer to TF NumPy Type Promotion for details):
tf.numpy.experimental_enable_numpy_behavior(dtype_conversion_mode="all")uses Jax type promotion rules.tf.numpy.experimental_enable_numpy_behavior(dtype_conversion_mode="safe")uses Jax type promotion rules, but disallows certain unsafe promotions.
NumPy Type Promotion
TensorFlow NumPy APIs have well-defined semantics for converting literals to ND array, as well as for performing type promotion on ND array inputs. Please see np.result_type for more details.
TensorFlow APIs leave tf.Tensor inputs unchanged and do not perform type promotion on them, while TensorFlow NumPy APIs promote all inputs according to NumPy type promotion rules. In the next example, you will perform type promotion. First, run addition on ND array inputs of different types and note the output types. None of these type promotions would be allowed by TensorFlow APIs.
print("Type promotion for operations")
values = [tnp.asarray(1, dtype=d) for d in
(tnp.int32, tnp.int64, tnp.float32, tnp.float64)]
for i, v1 in enumerate(values):
for v2 in values[i + 1:]:
print("%s + %s => %s" %
(v1.dtype.name, v2.dtype.name, (v1 + v2).dtype.name))
Type promotion for operations int32 + int64 => int64 int32 + float32 => float64 int32 + float64 => float64 int64 + float32 => float64 int64 + float64 => float64 float32 + float64 => float64
Finally, convert literals to ND array using ndarray.asarray and note the resulting type.
print("Type inference during array creation")
print("tnp.asarray(1).dtype == tnp.%s" % tnp.asarray(1).dtype.name)
print("tnp.asarray(1.).dtype == tnp.%s\n" % tnp.asarray(1.).dtype.name)
Type inference during array creation tnp.asarray(1).dtype == tnp.int64 tnp.asarray(1.).dtype == tnp.float64
When converting literals to ND array, NumPy prefers wide types like tnp.int64 and tnp.float64. In contrast, tf.convert_to_tensor prefers tf.int32 and tf.float32 types for converting constants to tf.Tensor. TensorFlow NumPy APIs adhere to the NumPy behavior for integers. As for floats, the prefer_float32 argument of experimental_enable_numpy_behavior lets you control whether to prefer tf.float32 over tf.float64 (default to False). For example:
tnp.experimental_enable_numpy_behavior(prefer_float32=True)
print("When prefer_float32 is True:")
print("tnp.asarray(1.).dtype == tnp.%s" % tnp.asarray(1.).dtype.name)
print("tnp.add(1., 2.).dtype == tnp.%s" % tnp.add(1., 2.).dtype.name)
tnp.experimental_enable_numpy_behavior(prefer_float32=False)
print("When prefer_float32 is False:")
print("tnp.asarray(1.).dtype == tnp.%s" % tnp.asarray(1.).dtype.name)
print("tnp.add(1., 2.).dtype == tnp.%s" % tnp.add(1., 2.).dtype.name)
When prefer_float32 is True: tnp.asarray(1.).dtype == tnp.float32 tnp.add(1., 2.).dtype == tnp.float32 When prefer_float32 is False: tnp.asarray(1.).dtype == tnp.float64 tnp.add(1., 2.).dtype == tnp.float64
Broadcasting
Similar to TensorFlow, NumPy defines rich semantics for "broadcasting" values. You can check out the NumPy broadcasting guide for more information and compare this with TensorFlow broadcasting semantics.
x = tnp.ones([2, 3])
y = tnp.ones([3])
z = tnp.ones([1, 2, 1])
print("Broadcasting shapes %s, %s and %s gives shape %s" % (
x.shape, y.shape, z.shape, (x + y + z).shape))
Broadcasting shapes (2, 3), (3,) and (1, 2, 1) gives shape (1, 2, 3)
Indexing
NumPy defines very sophisticated indexing rules. See the NumPy Indexing guide. Note the use of ND arrays as indices below.
x = tnp.arange(24).reshape(2, 3, 4)
print("Basic indexing")
print(x[1, tnp.newaxis, 1:3, ...], "\n")
print("Boolean indexing")
print(x[:, (True, False, True)], "\n")
print("Advanced indexing")
print(x[1, (0, 0, 1), tnp.asarray([0, 1, 1])])
Basic indexing tf.Tensor( [[[16 17 18 19] [20 21 22 23]]], shape=(1, 2, 4), dtype=int64) Boolean indexing tf.Tensor( [[[ 0 1 2 3] [ 8 9 10 11]] [[12 13 14 15] [20 21 22 23]]], shape=(2, 2, 4), dtype=int64) Advanced indexing tf.Tensor([12 13 17], shape=(3,), dtype=int64)
# Mutation is currently not supported
try:
tnp.arange(6)[1] = -1
except TypeError:
print("Currently, TensorFlow NumPy does not support mutation.")
Currently, TensorFlow NumPy does not support mutation.
Example Model
Next, you can see how to create a model and run inference on it. This simple model applies a relu layer followed by a linear projection. Later sections will show how to compute gradients for this model using TensorFlow's GradientTape.
class Model(object):
"""Model with a dense and a linear layer."""
def __init__(self):
self.weights = None
def predict(self, inputs):
if self.weights is None:
size = inputs.shape[1]
# Note that type `tnp.float32` is used for performance.
stddev = tnp.sqrt(size).astype(tnp.float32)
w1 = tnp.random.randn(size, 64).astype(tnp.float32) / stddev
bias = tnp.random.randn(64).astype(tnp.float32)
w2 = tnp.random.randn(64, 2).astype(tnp.float32) / 8
self.weights = (w1, bias, w2)
else:
w1, bias, w2 = self.weights
y = tnp.matmul(inputs, w1) + bias
y = tnp.maximum(y, 0) # Relu
return tnp.matmul(y, w2) # Linear projection
model = Model()
# Create input data and compute predictions.
print(model.predict(tnp.ones([2, 32], dtype=tnp.float32)))
tf.Tensor( [[-0.8292594 0.75780904] [-0.8292594 0.75780904]], shape=(2, 2), dtype=float32)
TensorFlow NumPy and NumPy
TensorFlow NumPy implements a subset of the full NumPy spec. While more symbols will be added over time, there are systematic features that will not be supported in the near future. These include NumPy C API support, Swig integration, Fortran storage order, views and stride_tricks, and some dtypes (like np.recarray and np.object). For more details, please see the TensorFlow NumPy API Documentation.
NumPy interoperability
TensorFlow ND arrays can interoperate with NumPy functions. These objects implement the __array__ interface. NumPy uses this interface to convert function arguments to np.ndarray values before processing them.
Similarly, TensorFlow NumPy functions can accept inputs of different types including np.ndarray. These inputs are converted to an ND array by calling ndarray.asarray on them.
Conversion of the ND array to and from np.ndarray may trigger actual data copies. Please see the section on buffer copies for more details.
# ND array passed into NumPy function.
np_sum = np.sum(tnp.ones([2, 3]))
print("sum = %s. Class: %s" % (float(np_sum), np_sum.__class__))
# `np.ndarray` passed into TensorFlow NumPy function.
tnp_sum = tnp.sum(np.ones([2, 3]))
print("sum = %s. Class: %s" % (float(tnp_sum), tnp_sum.__class__))
sum = 6.0. Class: <class 'numpy.float64'> sum = 6.0. Class: <class 'tensorflow.python.framework.ops.EagerTensor'>
# It is easy to plot ND arrays, given the __array__ interface.
labels = 15 + 2 * tnp.random.randn(1, 1000)
_ = plt.hist(labels)
Buffer copies
Intermixing TensorFlow NumPy with NumPy code may trigger data copies. This is because TensorFlow NumPy has stricter requirements on memory alignment than those of NumPy.
When a np.ndarray is passed to TensorFlow NumPy, it will check for alignment requirements and trigger a copy if needed. When passing an ND array CPU buffer to NumPy, generally the buffer will satisfy alignment requirements and NumPy will not need to create a copy.
ND arrays can refer to buffers placed on devices other than the local CPU memory. In such cases, invoking a NumPy function will trigger copies across the network or device as needed.
Given this, intermixing with NumPy API calls should generally be done with caution and the user should watch out for overheads of copying data. Interleaving TensorFlow NumPy calls with TensorFlow calls is generally safe and avoids copying data. See the section on TensorFlow interoperability for more details.
Operator precedence
TensorFlow NumPy defines an __array_priority__ higher than NumPy's. This means that for operators involving both ND array and np.ndarray, the former will take precedence, i.e., np.ndarray input will get converted to an ND array and the TensorFlow NumPy implementation of the operator will get invoked.
x = tnp.ones([2]) + np.ones([2])
print("x = %s\nclass = %s" % (x, x.__class__))
x = tf.Tensor([2. 2.], shape=(2,), dtype=float64) class = <class 'tensorflow.python.framework.ops.EagerTensor'>
TF NumPy and TensorFlow
TensorFlow NumPy is built on top of TensorFlow and hence interoperates seamlessly with TensorFlow.
tf.Tensor and ND array
ND array is an alias to tf.Tensor, so obviously they can be intermixed without triggering actual data copies.
x = tf.constant([1, 2])
print(x)
# `asarray` and `convert_to_tensor` here are no-ops.
tnp_x = tnp.asarray(x)
print(tnp_x)
print(tf.convert_to_tensor(tnp_x))
# Note that tf.Tensor.numpy() will continue to return `np.ndarray`.
print(x.numpy(), x.numpy().__class__)
tf.Tensor([1 2], shape=(2,), dtype=int32) tf.Tensor([1 2], shape=(2,), dtype=int32) tf.Tensor([1 2], shape=(2,), dtype=int32) [1 2] <class 'numpy.ndarray'>
TensorFlow interoperability
An ND array can be passed to TensorFlow APIs, since ND array is just an alias to tf.Tensor. As mentioned earlier, such interoperation does not do data copies, even for data placed on accelerators or remote devices.
Conversely, tf.Tensor objects can be passed to tf.experimental.numpy APIs, without performing data copies.
# ND array passed into TensorFlow function.
tf_sum = tf.reduce_sum(tnp.ones([2, 3], tnp.float32))
print("Output = %s" % tf_sum)
# `tf.Tensor` passed into TensorFlow NumPy function.
tnp_sum = tnp.sum(tf.ones([2, 3]))
print("Output = %s" % tnp_sum)
Output = tf.Tensor(6.0, shape=(), dtype=float32) Output = tf.Tensor(6.0, shape=(), dtype=float32)
Gradients and Jacobians: tf.GradientTape
TensorFlow's GradientTape can be used for backpropagation through TensorFlow and TensorFlow NumPy code.
Use the model created in Example Model section, and compute gradients and jacobians.
def create_batch(batch_size=32):
"""Creates a batch of input and labels."""
return (tnp.random.randn(batch_size, 32).astype(tnp.float32),
tnp.random.randn(batch_size, 2).astype(tnp.float32))
def compute_gradients(model, inputs, labels):
"""Computes gradients of squared loss between model prediction and labels."""
with tf.GradientTape() as tape:
assert model.weights is not None
# Note that `model.weights` need to be explicitly watched since they
# are not tf.Variables.
tape.watch(model.weights)
# Compute prediction and loss
prediction = model.predict(inputs)
loss = tnp.sum(tnp.square(prediction - labels))
# This call computes the gradient through the computation above.
return tape.gradient(loss, model.weights)
inputs, labels = create_batch()
gradients = compute_gradients(model, inputs, labels)
# Inspect the shapes of returned gradients to verify they match the
# parameter shapes.
print("Parameter shapes:", [w.shape for w in model.weights])
print("Gradient shapes:", [g.shape for g in gradients])
# Verify that gradients are of type ND array.
assert isinstance(gradients[0], tnp.ndarray)
Parameter shapes: [TensorShape([32, 64]), TensorShape([64]), TensorShape([64, 2])] Gradient shapes: [TensorShape([32, 64]), TensorShape([64]), TensorShape([64, 2])]
# Computes a batch of jacobians. Each row is the jacobian of an element in the
# batch of outputs w.r.t. the corresponding input batch element.
def prediction_batch_jacobian(inputs):
with tf.GradientTape() as tape:
tape.watch(inputs)
prediction = model.predict(inputs)
return prediction, tape.batch_jacobian(prediction, inputs)
inp_batch = tnp.ones([16, 32], tnp.float32)
output, batch_jacobian = prediction_batch_jacobian(inp_batch)
# Note how the batch jacobian shape relates to the input and output shapes.
print("Output shape: %s, input shape: %s" % (output.shape, inp_batch.shape))
print("Batch jacobian shape:", batch_jacobian.shape)
Output shape: (16, 2), input shape: (16, 32) Batch jacobian shape: (16, 2, 32)
Trace compilation: tf.function
TensorFlow's tf.function works by "trace compiling" the code and then optimizing these traces for much faster performance. See the Introduction to Graphs and Functions.
tf.function can be used to optimize TensorFlow NumPy code as well. Here is a simple example to demonstrate the speedups. Note that the body of tf.function code includes calls to TensorFlow NumPy APIs.
inputs, labels = create_batch(512)
print("Eager performance")
compute_gradients(model, inputs, labels)
print(timeit.timeit(lambda: compute_gradients(model, inputs, labels),
number=10) * 100, "ms")
print("\ntf.function compiled performance")
compiled_compute_gradients = tf.function(compute_gradients)
compiled_compute_gradients(model, inputs, labels) # warmup
print(timeit.timeit(lambda: compiled_compute_gradients(model, inputs, labels),
number=10) * 100, "ms")
Eager performance 2.710705300000882 ms tf.function compiled performance 0.7041131000050882 ms
Vectorization: tf.vectorized_map
TensorFlow has inbuilt support for vectorizing parallel loops, which allows speedups of one to two orders of magnitude. These speedups are accessible via the tf.vectorized_map API and apply to TensorFlow NumPy code as well.
It is sometimes useful to compute the gradient of each output in a batch w.r.t. the corresponding input batch element. Such computation can be done efficiently using tf.vectorized_map as shown below.
@tf.function
def vectorized_per_example_gradients(inputs, labels):
def single_example_gradient(arg):
inp, label = arg
return compute_gradients(model,
tnp.expand_dims(inp, 0),
tnp.expand_dims(label, 0))
# Note that a call to `tf.vectorized_map` semantically maps
# `single_example_gradient` over each row of `inputs` and `labels`.
# The interface is similar to `tf.map_fn`.
# The underlying machinery vectorizes away this map loop which gives
# nice speedups.
return tf.vectorized_map(single_example_gradient, (inputs, labels))
batch_size = 128
inputs, labels = create_batch(batch_size)
per_example_gradients = vectorized_per_example_gradients(inputs, labels)
for w, p in zip(model.weights, per_example_gradients):
print("Weight shape: %s, batch size: %s, per example gradient shape: %s " % (
w.shape, batch_size, p.shape))
Weight shape: (32, 64), batch size: 128, per example gradient shape: (128, 32, 64) Weight shape: (64,), batch size: 128, per example gradient shape: (128, 64) Weight shape: (64, 2), batch size: 128, per example gradient shape: (128, 64, 2)
# Benchmark the vectorized computation above and compare with
# unvectorized sequential computation using `tf.map_fn`.
@tf.function
def unvectorized_per_example_gradients(inputs, labels):
def single_example_gradient(arg):
inp, label = arg
return compute_gradients(model,
tnp.expand_dims(inp, 0),
tnp.expand_dims(label, 0))
return tf.map_fn(single_example_gradient, (inputs, labels),
fn_output_signature=(tf.float32, tf.float32, tf.float32))
print("Running vectorized computation")
print(timeit.timeit(lambda: vectorized_per_example_gradients(inputs, labels),
number=10) * 100, "ms")
print("\nRunning unvectorized computation")
per_example_gradients = unvectorized_per_example_gradients(inputs, labels)
print(timeit.timeit(lambda: unvectorized_per_example_gradients(inputs, labels),
number=10) * 100, "ms")
Running vectorized computation 0.659675699989748 ms Running unvectorized computation 29.259711299982882 ms
Device placement
TensorFlow NumPy can place operations on CPUs, GPUs, TPUs and remote devices. It uses standard TensorFlow mechanisms for device placement. Below a simple example shows how to list all devices and then place some computation on a particular device.
TensorFlow also has APIs for replicating computation across devices and performing collective reductions which will not be covered here.
List devices
tf.config.list_logical_devices and tf.config.list_physical_devices can be used to find what devices to use.
print("All logical devices:", tf.config.list_logical_devices())
print("All physical devices:", tf.config.list_physical_devices())
# Try to get the GPU device. If unavailable, fallback to CPU.
try:
device = tf.config.list_logical_devices(device_type="GPU")[0]
except IndexError:
device = "/device:CPU:0"
All logical devices: [LogicalDevice(name='/device:CPU:0', device_type='CPU'), LogicalDevice(name='/device:GPU:0', device_type='GPU'), LogicalDevice(name='/device:GPU:1', device_type='GPU'), LogicalDevice(name='/device:GPU:2', device_type='GPU'), LogicalDevice(name='/device:GPU:3', device_type='GPU')] All physical devices: [PhysicalDevice(name='/physical_device:CPU:0', device_type='CPU'), PhysicalDevice(name='/physical_device:GPU:0', device_type='GPU'), PhysicalDevice(name='/physical_device:GPU:1', device_type='GPU'), PhysicalDevice(name='/physical_device:GPU:2', device_type='GPU'), PhysicalDevice(name='/physical_device:GPU:3', device_type='GPU')]
Placing operations: tf.device
Operations can be placed on a device by calling it in a tf.device scope.
print("Using device: %s" % str(device))
# Run operations in the `tf.device` scope.
# If a GPU is available, these operations execute on the GPU and outputs are
# placed on the GPU memory.
with tf.device(device):
prediction = model.predict(create_batch(5)[0])
print("prediction is placed on %s" % prediction.device)
Using device: LogicalDevice(name='/device:GPU:0', device_type='GPU') prediction is placed on /job:localhost/replica:0/task:0/device:GPU:0
Copying ND arrays across devices: tnp.copy
A call to tnp.copy, placed in a certain device scope, will copy the data to that device, unless the data is already on that device.
with tf.device("/device:CPU:0"):
prediction_cpu = tnp.copy(prediction)
print(prediction.device)
print(prediction_cpu.device)
/job:localhost/replica:0/task:0/device:GPU:0 /job:localhost/replica:0/task:0/device:CPU:0
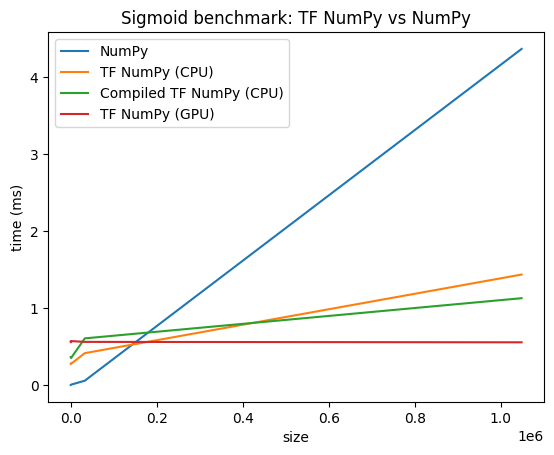
Performance comparisons
TensorFlow NumPy uses highly optimized TensorFlow kernels that can be dispatched on CPUs, GPUs and TPUs. TensorFlow also performs many compiler optimizations, like operation fusion, which translate to performance and memory improvements. See TensorFlow graph optimization with Grappler to learn more.
However TensorFlow has higher overheads for dispatching operations compared to NumPy. For workloads composed of small operations (less than about 10 microseconds), these overheads can dominate the runtime and NumPy could provide better performance. For other cases, TensorFlow should generally provide better performance.
Run the benchmark below to compare NumPy and TensorFlow NumPy performance for different input sizes.
def benchmark(f, inputs, number=30, force_gpu_sync=False):
"""Utility to benchmark `f` on each value in `inputs`."""
times = []
for inp in inputs:
def _g():
if force_gpu_sync:
one = tnp.asarray(1)
f(inp)
if force_gpu_sync:
with tf.device("CPU:0"):
tnp.copy(one) # Force a sync for GPU case
_g() # warmup
t = timeit.timeit(_g, number=number)
times.append(t * 1000. / number)
return times
def plot(np_times, tnp_times, compiled_tnp_times, has_gpu, tnp_times_gpu):
"""Plot the different runtimes."""
plt.xlabel("size")
plt.ylabel("time (ms)")
plt.title("Sigmoid benchmark: TF NumPy vs NumPy")
plt.plot(sizes, np_times, label="NumPy")
plt.plot(sizes, tnp_times, label="TF NumPy (CPU)")
plt.plot(sizes, compiled_tnp_times, label="Compiled TF NumPy (CPU)")
if has_gpu:
plt.plot(sizes, tnp_times_gpu, label="TF NumPy (GPU)")
plt.legend()
# Define a simple implementation of `sigmoid`, and benchmark it using
# NumPy and TensorFlow NumPy for different input sizes.
def np_sigmoid(y):
return 1. / (1. + np.exp(-y))
def tnp_sigmoid(y):
return 1. / (1. + tnp.exp(-y))
@tf.function
def compiled_tnp_sigmoid(y):
return tnp_sigmoid(y)
sizes = (2 ** 0, 2 ** 5, 2 ** 10, 2 ** 15, 2 ** 20)
np_inputs = [np.random.randn(size).astype(np.float32) for size in sizes]
np_times = benchmark(np_sigmoid, np_inputs)
with tf.device("/device:CPU:0"):
tnp_inputs = [tnp.random.randn(size).astype(np.float32) for size in sizes]
tnp_times = benchmark(tnp_sigmoid, tnp_inputs)
compiled_tnp_times = benchmark(compiled_tnp_sigmoid, tnp_inputs)
has_gpu = len(tf.config.list_logical_devices("GPU"))
if has_gpu:
with tf.device("/device:GPU:0"):
tnp_inputs = [tnp.random.randn(size).astype(np.float32) for size in sizes]
tnp_times_gpu = benchmark(compiled_tnp_sigmoid, tnp_inputs, 100, True)
else:
tnp_times_gpu = None
plot(np_times, tnp_times, compiled_tnp_times, has_gpu, tnp_times_gpu)